Recently, the research team led by Dr. Wanlu Liu at the Zhejiang University–University of Edinburgh Institute, together with Prof. Linrong Lu’s group at the Zhejiang University School of Medicine, in collaboration with the Tencent AI for Life Sciences Lab and several other institutions, published a research article entitled “A pan-disease and population-level single-cell TCRαβ repertoire reference” in the international journal Cell Discovery. This study presents, for the first time, a comprehensive single-cell T cell receptor (TCRαβ) reference atlas that spans multiple populations and disease conditions, and introduces an innovative computational framework, TCR-DeepInsight, providing a new tool and resource for systematically exploring the relationship between TCRs and human diseases.

T cells play a pivotal role in adaptive immunity by recognizing pathogen- or tumor-derived peptides presented by major histocompatibility complex (MHC) molecules through their surface T cell receptors (TCRs). Although TCR sequences are highly diverse due to random V(D)J recombination, identical or highly similar “public TCRs” have been observed across different individuals, often reflecting shared immune responses to common pathogens or disease-associated antigens.
With the advent of single-cell RNA/TCR sequencing, it has become possible to simultaneously capture TCR sequences and transcriptomic profiles from the same cells. However, a comprehensive integration of these data across populations and diseases has remained a major challenge in immunology.
Key Findings
1. Construction of the Largest Single-Cell TCRαβ Reference Database
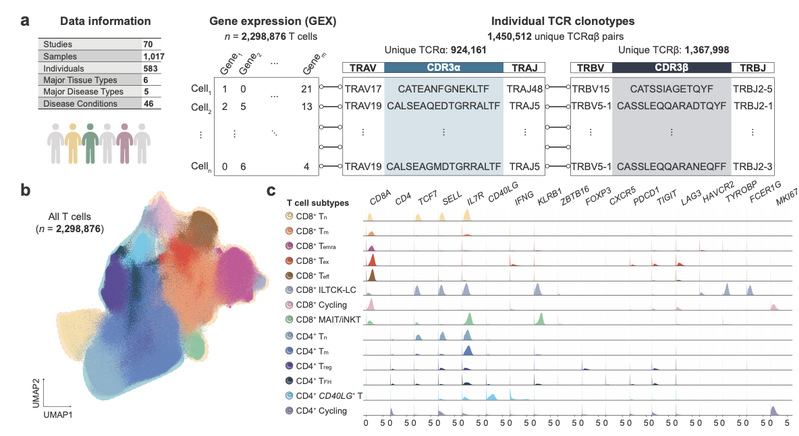
The team collected and integrated data from 70 studies, covering 1017 biological samples, 583 individuals, and 46 disease conditions, including cancers, autoimmune disorders, infections, and healthy controls.
In total, the atlas encompasses over 2.29 million high-confidence T cells with matched full-length TCRαβ sequences, transcriptomes, and HLA genotypes, now publicly available through the huARdb 2.0 database.
2. Revealing Intrinsic Genetic Determinants of T Cell Lineage Commitment
By analyzing the pairing of V/J genes and amino acid usage across CD4⁺ and CD8⁺ T cells, the study uncovered intrinsic TCR sequence features that underlie MHC class I or class II restriction, providing new insights into T cell lineage specification.
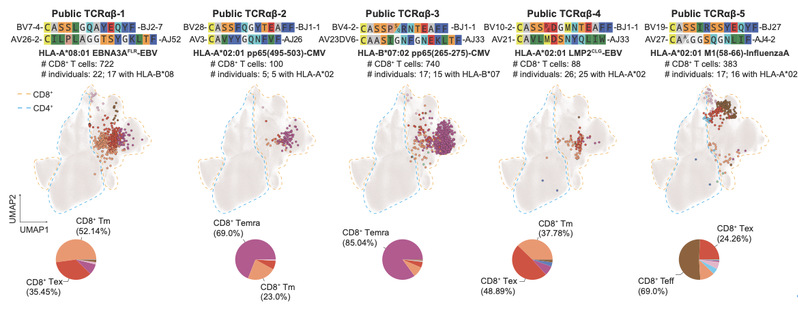
3. Identification of Public TCRs Shared Across Populations
The researchers discovered nearly 3,000 public TCRαβ clonotypes, which are more clonally expanded and frequently associated with shared HLA alleles. Many of these public receptors are predicted to recognize epitopes derived from common viruses such as Epstein-Barr virus (EBV), cytomegalovirus (CMV), and influenza A virus, revealing conserved patterns of immune defense across individuals.
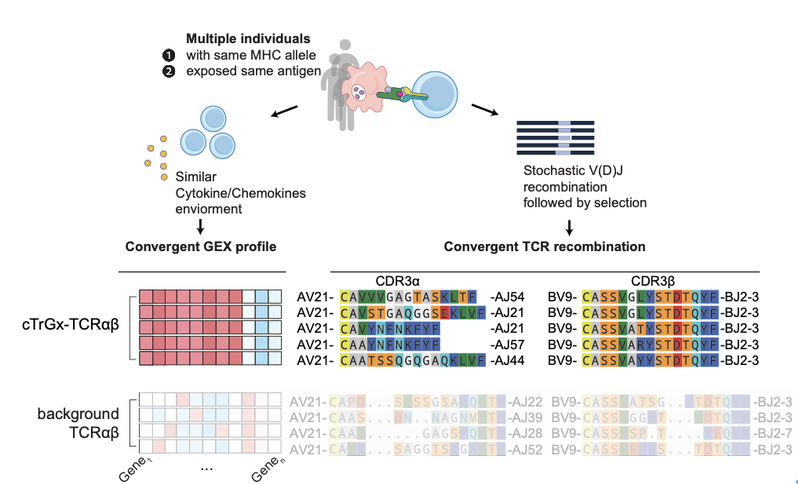
4. Development of the TCR-DeepInsight Computational Framework
The team introduced TCR-DeepInsight, a deep learning–based approach that integrates TCR sequence and transcriptomic information to identify HLA-shared and disease-associated TCR clusters.
Leveraging large-scale representation learning (via BERT and variational autoencoders), this framework enables researchers to map their own datasets to the reference atlas for cross-population and pan-disease comparison of functional TCR repertoires.
This work establishes an unprecedented pan-disease and population-levelsingle-cell TCR reference that bridges large-scale immunogenomic data with advanced computational modeling.
The study not only deepens our understanding of TCR diversity, lineage restriction, and immune memory formation, but also lays a foundation for precision immunotherapy, vaccine design, and immune repertoire–based diagnostics.
Moving forward, the research team aims to expand the atlas to encompass additional populations and disease types, and to integrate artificial intelligence (AI)–based antigen prediction models to advance precision immunology.
Dr. Wanlu Liu from the Zhejiang University–University of Edinburgh Institute (ZJE) and Prof. Linrong Lu from the Zhejiang University School of Medicine are the corresponding authors of this paper. Ziwei Xue, a dual Ph.D. student of the ZJE Joint Doctoral Program, Lize Wu, a Ph.D. candidate at the Zhejiang University School of Medicine, and Dr. Bing Gao graduated from ZJE are co–first authors. This research was conducted in close collaboration with the Tencent AI for Life Sciences Lab. In addition, Yiru Chen, Yicheng Qi, and Tianze Dong, undergraduate students majoring in Bioinformatics (Class of 2022) at ZJE, also participated in this study.







